| PCR Colored 5xReaction Buffer |
| GemTaq™ Hot Start DNA Polymerase |
GemTaq™ DNA Polymerase |
NEW PRODUCT SNPase Hot Start DNA Polymerase |

| MGQuest |
| 2007-2014 MGQuest. All rights reserved. |
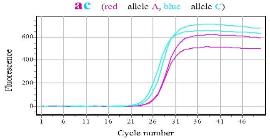
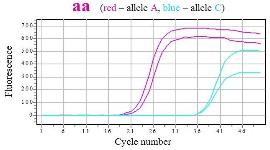
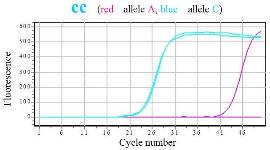
SNPase Hot Start in allele specific PCR Gene MTHFR1298 (a→c), qPCR mix with SYBR green |
SNPase Hot Start and KlenThermase belong to the same family of high fidelity thermostable DNA polymerases. Proprietary amino acid
substitutions introduced into the active center of the enzymes cause dramatic increase of specificity and fidelity of dNTPs, ddNTPs
incorporation that is 10-15 times higher than a regular Taq DNA Polymerase. SNPase Hot Start and KlenThermase were designed for a
single-nucleotide polymorphism (SNP) detection and cycle sequencing with ddNTPs, respectively. These enzymes have only 5'-3'
polymerase activity and lack exonuclease activity. SNPase Hot Start has a modified polymerase activity that is restored during the initial
denaturation step allowing for a hot-start PCR and eliminates the presence of non-specific products.
SNPase and KlenThermase were recognized as one of the best polymerases in SNP detection by allele-specific
PCR and mini-sequencing.
substitutions introduced into the active center of the enzymes cause dramatic increase of specificity and fidelity of dNTPs, ddNTPs
incorporation that is 10-15 times higher than a regular Taq DNA Polymerase. SNPase Hot Start and KlenThermase were designed for a
single-nucleotide polymorphism (SNP) detection and cycle sequencing with ddNTPs, respectively. These enzymes have only 5'-3'
polymerase activity and lack exonuclease activity. SNPase Hot Start has a modified polymerase activity that is restored during the initial
denaturation step allowing for a hot-start PCR and eliminates the presence of non-specific products.
SNPase and KlenThermase were recognized as one of the best polymerases in SNP detection by allele-specific
PCR and mini-sequencing.
Advantages
- High sensitivity to mismatches at 3’-end of primers
- High fidelity of dNTPs and ddNTPs incorporation
- Lack of 5’-3’ and 3’-5’exonuclease activity
- High specificity, low level of background PCR
- Fast activation (5-10 sec, 95°C)
- TA cloning
Applications
- SNP- genotyping, allele specific PCR, allele specific primer extension
- High fidelity PCR (up to 1000 bp)
- Multiplex PCR
- Real Time PCR with intercalating dyes (SYBR Green, Eva Green)
- Mini-sequencing
GemTaq™ DNA Polymerase is a special formulation of standard Taq DNA Polymerase that features significantly improved amplifications
of DNA fragments. GemTaq™ DNA Polymerase has an elongation rate higher than regular Taq DNA Polymerase, resulting in robust yields
and greater sensitivity of PCR amplifications within a short reaction time.
of DNA fragments. GemTaq™ DNA Polymerase has an elongation rate higher than regular Taq DNA Polymerase, resulting in robust yields
and greater sensitivity of PCR amplifications within a short reaction time.

B
A
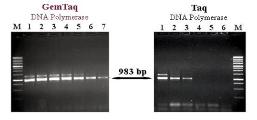
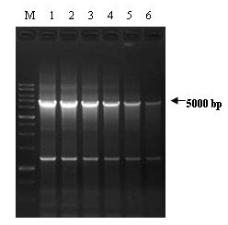
Fig.4. PCR amplifications performed with GemTaq™ DNA Polymerase in the Colored Reaction Buffer.
Lambda DNA fragment (5000 bp) was amplified in 50 µl with 2U (lane 1), 1.25U (lane 2), 0.83U (lane 3), 0.55U (lane 4), 0.37U (lane 5)
and 0.25U (lane 6) of GemTaq™ DNA Polymerase in the Colored Reaction Buffer. Each PCR reaction was performed from 2 ng of Lambda
DNA as a template during 30 cycles. Lane M: 1 kb DNA ladder.
Lambda DNA fragment (5000 bp) was amplified in 50 µl with 2U (lane 1), 1.25U (lane 2), 0.83U (lane 3), 0.55U (lane 4), 0.37U (lane 5)
and 0.25U (lane 6) of GemTaq™ DNA Polymerase in the Colored Reaction Buffer. Each PCR reaction was performed from 2 ng of Lambda
DNA as a template during 30 cycles. Lane M: 1 kb DNA ladder.
The PCR Colored 5xReaction Buffer is supplied with GemTaq™ DNA
Polymerase (Cat #EP022, #EP023, #EP024, #EP025) and GemTaq™ Hot
Start DNA Polymerase (Cat #EP042, #EP043, #EP044, #EP045). It is
also part of GemTaq™ Colored 5xMaster Mix (Cat #EPM21, #EPM22,
#EPM23, #EPM24) and GemTaq™ Hot Start Colored 5xMaster Mix
(Cat #EPM41, #EPM42, #EPM43, #EPM44).
Polymerase (Cat #EP022, #EP023, #EP024, #EP025) and GemTaq™ Hot
Start DNA Polymerase (Cat #EP042, #EP043, #EP044, #EP045). It is
also part of GemTaq™ Colored 5xMaster Mix (Cat #EPM21, #EPM22,
#EPM23, #EPM24) and GemTaq™ Hot Start Colored 5xMaster Mix
(Cat #EPM41, #EPM42, #EPM43, #EPM44).
to add a loading sample buffer. It contains a compound that increases sample density for electrophoresis and two dyes (Red and Yellow)
that separate during electrophoresis to allow for easy monitoring. The presence of the dyes has no effect on most PCR amplifications
(Fig.4A). The Red and Yellow dyes migrate at the same rate as 800-1000 bp DNA fragments and 20-30 bp DNA fragments in a 1% agarose
gel, respectively (Fig.4B).
that separate during electrophoresis to allow for easy monitoring. The presence of the dyes has no effect on most PCR amplifications
(Fig.4A). The Red and Yellow dyes migrate at the same rate as 800-1000 bp DNA fragments and 20-30 bp DNA fragments in a 1% agarose
gel, respectively (Fig.4B).
Applications
- Hot start PCR.
- Low-copy target.
- High throughput PCR applications
- Complex genomic templates.
- TA cloning.
Advantages
- Ultra-high specificity.
- Fast activation.
- Greater efficiency and sensitivity in a wide range of PCR
assays within a short reaction time. - Recommended for low copy assays.
- Amplifies fragments up to 5 kb.
- Cost-effective.
GemTaq™ Hot Start DNA Polymerase is a heat-activated high-performance proprietary formulation of GemTaq™ DNA Polymerase.The
modified polymerase activity is restored during the initial denaturation step (94-95 °C for 3 min.) allowing for a hot-start PCR. Therefore
GemTaq™ Hot Start DNA Polymerase provides improved specificity and very high PCR sensitivity by eliminating the presence of non-
specifics products. It has an elongation rate higher than regular Taq DNA Polymerase, resulting in robust yields and greater sensitivity of
PCR amplifications within a short reaction time.
modified polymerase activity is restored during the initial denaturation step (94-95 °C for 3 min.) allowing for a hot-start PCR. Therefore
GemTaq™ Hot Start DNA Polymerase provides improved specificity and very high PCR sensitivity by eliminating the presence of non-
specifics products. It has an elongation rate higher than regular Taq DNA Polymerase, resulting in robust yields and greater sensitivity of
PCR amplifications within a short reaction time.
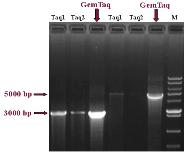
Fig.3. Greater efficiency of GemTaq™ DNA Polymerase in PCR
amplifications.
Lambda DNA fragments (3000 bp and 5000 bp) were amplified in 50 µl with 0.5 U of
GemTaq™ DNA Polymerase and standard Taq DNA polymerases in supplied Reaction
Buffers respectively. Each PCR reaction was performed from 5 ng of Lambda DNA as a
template during 28 cycles.
amplifications.
Lambda DNA fragments (3000 bp and 5000 bp) were amplified in 50 µl with 0.5 U of
GemTaq™ DNA Polymerase and standard Taq DNA polymerases in supplied Reaction
Buffers respectively. Each PCR reaction was performed from 5 ng of Lambda DNA as a
template during 28 cycles.
Fig.2. Greater sensitivity of GemTaq™ DNA Polymerase in PCR
amplifications.
The β-Actin gene fragment (850 bp) was amplified in 25 µl from the indicated amounts of
human genomic DNA as a template. Each PCR reaction was performed using 0.25 U of
GemTaq™ DNA Polymerase and standard Taq DNA Polymerases (Taq 1 from competitor 1
Taq 2 from competitor 2) in supplied Reaction Buffers respectively during 33 cycles.
amplifications.
The β-Actin gene fragment (850 bp) was amplified in 25 µl from the indicated amounts of
human genomic DNA as a template. Each PCR reaction was performed using 0.25 U of
GemTaq™ DNA Polymerase and standard Taq DNA Polymerases (Taq 1 from competitor 1
Taq 2 from competitor 2) in supplied Reaction Buffers respectively during 33 cycles.
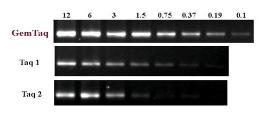
Fig.1 Higher efficiency of GemTaq™ DNA Polymerase in PCR
amplifications.
The G3PDH gene fragment (983 bp) was amplified in 50 µl with different amounts of
GemTaq™ DNA Polymerase and standard Taq DNA Polymerase (line1 - 2.5 U, line2 –
1.25 U, line3 – 0.62 U, line4 – 0.31 U, line5 – 0.15 U, line6 – 0.075 U, line7 – 0.037 U) in
supplied Reaction Buffers respectively. Each PCR reaction was performed from 50 ng of
human genomic DNA as a template during 28 cycles.
amplifications.
The G3PDH gene fragment (983 bp) was amplified in 50 µl with different amounts of
GemTaq™ DNA Polymerase and standard Taq DNA Polymerase (line1 - 2.5 U, line2 –
1.25 U, line3 – 0.62 U, line4 – 0.31 U, line5 – 0.15 U, line6 – 0.075 U, line7 – 0.037 U) in
supplied Reaction Buffers respectively. Each PCR reaction was performed from 50 ng of
human genomic DNA as a template during 28 cycles.
Advantages
DNA Polymerase
Applications
- Higher efficiency (Fig.1) and greater sensitivity (Fig 2) in
DNA Polymerase
- Suited for amplifications of long DNA fragments (Fig.3)
- Shorter reaction times due to higher elongation rate
- Unbeatable value in terms of cost per unit and
performance
Applications
- Routine and high-throughput PCR
- Primer extension
- DNA microarray analysis
- TA cloning
| Certified WBE (Women's Business Enterprise) |
Product Citations
- Lovmar, L., Ahlford, A., Jonsson, M. and Syvänen, A.-C. (2005)
Silhouette scores for assessment of SNP genotype clusters. BMC
Genomics, 6:35
- Schnorrer, F., Ahlford, A., Chen, D., Milani, L. and Syvänen, A.-C.
(2008) Positional cloning by fast-track SNP-mapping in Drosophila
melanogaster. Nature Protocols 3, 1751 - 1765
- Ahlford, A., Kjeldsen, B., Reimers, J., Lundmark, A., Romani, M.,
Wolff, A., Syvänen, A.-C. and Brivio, M. (2010) Dried reagents for
multiplex genotyping by tag-array minisequencing to be used in
microfluidic devices. Analyst,135, 2377-2385
techserv@mgquest.com
sales@mgquest.com
sales@mgquest.com